Welcome to the web pages of the Radford Group
Our research
One of the most fascinating questions in biology is how proteins are able to fold and assemble into complex, functional entities given just the information provided by the amino acid sequence. A related, equally important facet of the same fundamental question is how protein misfolding can lead to protein aggregation, cellular dysfunction and disease. These issues are the major focus of my research and are tackled using a broad range of techniques including protein chemistry, structural molecular biology, chemical biology, cell biology and biophysical methods.
Current major projects:
- Mechanism(s) of protein misfolding and assembly into amyloid
- Outer Membrane Protein (OMP) folding – The role of chaperones & BAM
- Stabilising proteins of therapeutic interest against aggregation
- Method development (MS, NMR, single molecule, biophysical methods)
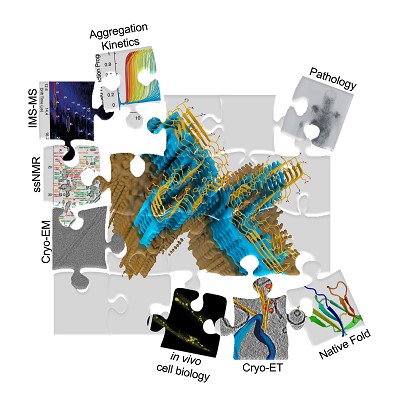
How and why proteins aggregate into amyloid are important fundamental questions that have far-reaching biomedical importance. Focusing on the proteins (β2-microglobulin (dialysis related amyloidosis)); amylin (type II diabetes); α-synuclein (Parkinson’s) and Aβ (Alzheimer’s), work in the group aims to map the structural mechanism of amyloid formation and to develop reagents to control aggregation in vitro and in vivo. Recent highlights include structure determination of oligomers (NMR (Karamanos (2019) eLife)) and fibrils (cryoEM (with Ranson) (Gallardo (2020) Nature Struct. Mol. Biol.; Wilkinson (2023) Nature Comms.)); discovery of small molecules that modulate amyloid formation (Cawood (2020) JACS; Xu (2022) Nature Comms.) and demonstration that early protein-protein interactions in amyloid formation are specific and can be targeted to arrest assembly in vitro and in vivo (Doherty (2020) Nature Struct. Mol. Biol.; Ulamec (2022) Nature Comms, Guthertz (2022) PNAS).
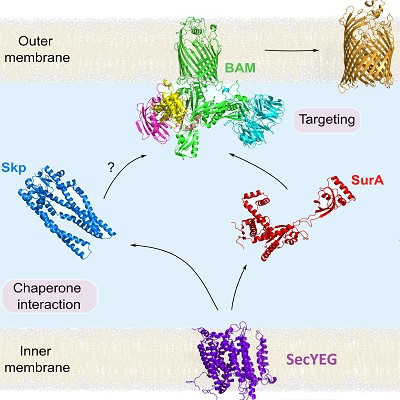
How OMPs fold and assemble into the asymmetric outer-membrane (OM) of Gram-negative bacteria is a second research theme in our lab. In a collaborative multidisciplinary team (with David Brockwell, Neil Ranson, Roman Tuma and Ian Collinson (Bristol)) we are investigating how OMPs cross the inner-membrane via SecYEG (Fessl (2018) eLife)); traverse the periplasm aided by chaperones (Skp/SurA) (Calabrese (2020) Nature Comms)), fold into membranes in vitro (Vorobieva (2021) Science) and in vivo catalysed by the essential β-barrel membrane machinery (BAM) (Iadanza (2020) Comms. Biol.; Schriffrin (2022) Comms. Biol.). Building on these insights we are currently exploring the dynamic motions of BAM during catalysis and how this can be harnessed to generate new antibacterial agents against Gram-negative pathogens (White (2021) Nature Comms.).
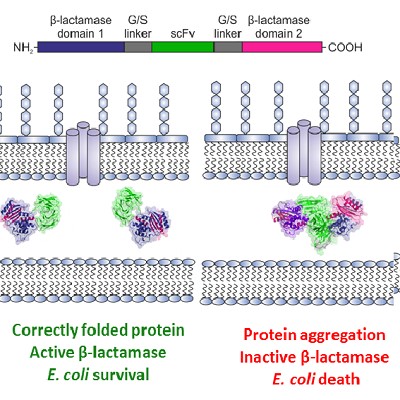
We are also exploiting our knowledge of protein folding/aggregation to practical benefit by screening amyloidogenic proteins, as well as proteins of interest to biopharma, for hotspots that cause aggregation. By coupling aggregation to bacterial growth using a tripartite β-lactamase fusion construct we have discovered small molecules that prevent aggregation of amyloidogenic proteins (Saunders (2016) Nature Chem. Biol.). With David Brockwell and our collaborators in AstraZeneca, we recently combined the assay with directed evolution to enhance the resilience of biopharmaceutically-relevant antibodies to aggregation (Ebo (2020) Nature Comms.). Finally, in collaboration with David Brockwell and Nik Kapur (Mechanical Engineering, Leeds), we are examining how flow fields enhance, or cause, aggregation by flow-induced protein deformations (Dobson (2017) PNAS, Willis (2020) Eng. Rep.).
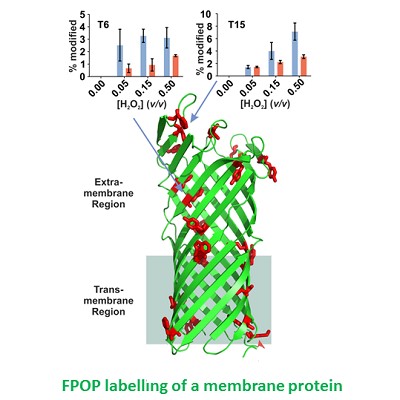
Major developments in methods and instrumentation have played a key role in increasing in our understanding of protein folding and aggregation. Future developments in these fields will also require innovative approaches that cross the boundaries between disciplines. We have been involved in many exciting collaborations to fulfil this aim. With Alison Ashcroft (now emeritus) and Frank Sobott we have developed and expanded our arsenal of methods to interrogate protein folding, protein-protein interactions and protein complexes, including MS methods (HDX-MS and fast photochemical oxidation of proteins (FPOP-MS)) (Cornwell (2018) J. Am. Soc. MS, Cornwell (2019) Analyt. Chem.); ion mobility MS to determine the effect of ligand binding on amyloidogenic monomers and oligomers (Young (2023) J. Am. Soc. MS.) and, with Andrew Wilson (Chemistry) we have developed rapid crosslinking to map protein-chaperone interactions (Horne (2018) Angewandte Chemie, Calabrese (2020) Nature Comms). Developments in NMR methods remain a mainstay of our laboratory activities (Karamanos (2022) Frontiers Neurosci.). In collaboration with David Brockwell and George Heath we are involved in exciting developments in the use of the AFM for measurements of protein unfolding and protein binding (Ulamec (2021) Nature Comms.) and with Paolo Actis (Mechanical Engineering) in the development of nanopores for the manipulation and identification of protein assemblies (Chau (2020) Nanoletters; Chau (2022) ACS Nano.).